MRIPredict
Free R package to easily predict diagnosis from sMRI scans
Created by the Imaging of mood- and anxiety- related disorders (IMARD) group,
IDIBAPS, Hospital Clínic de Barcelona
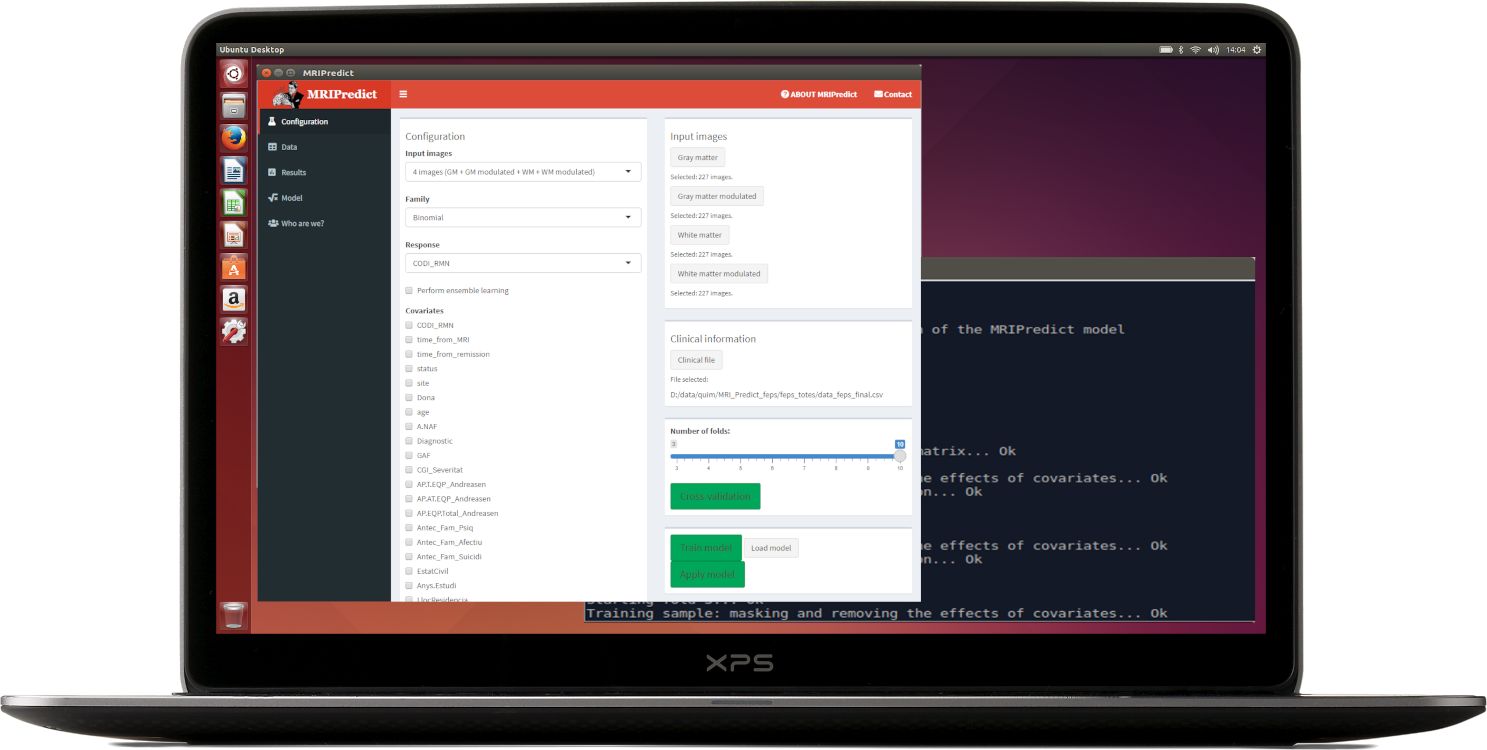
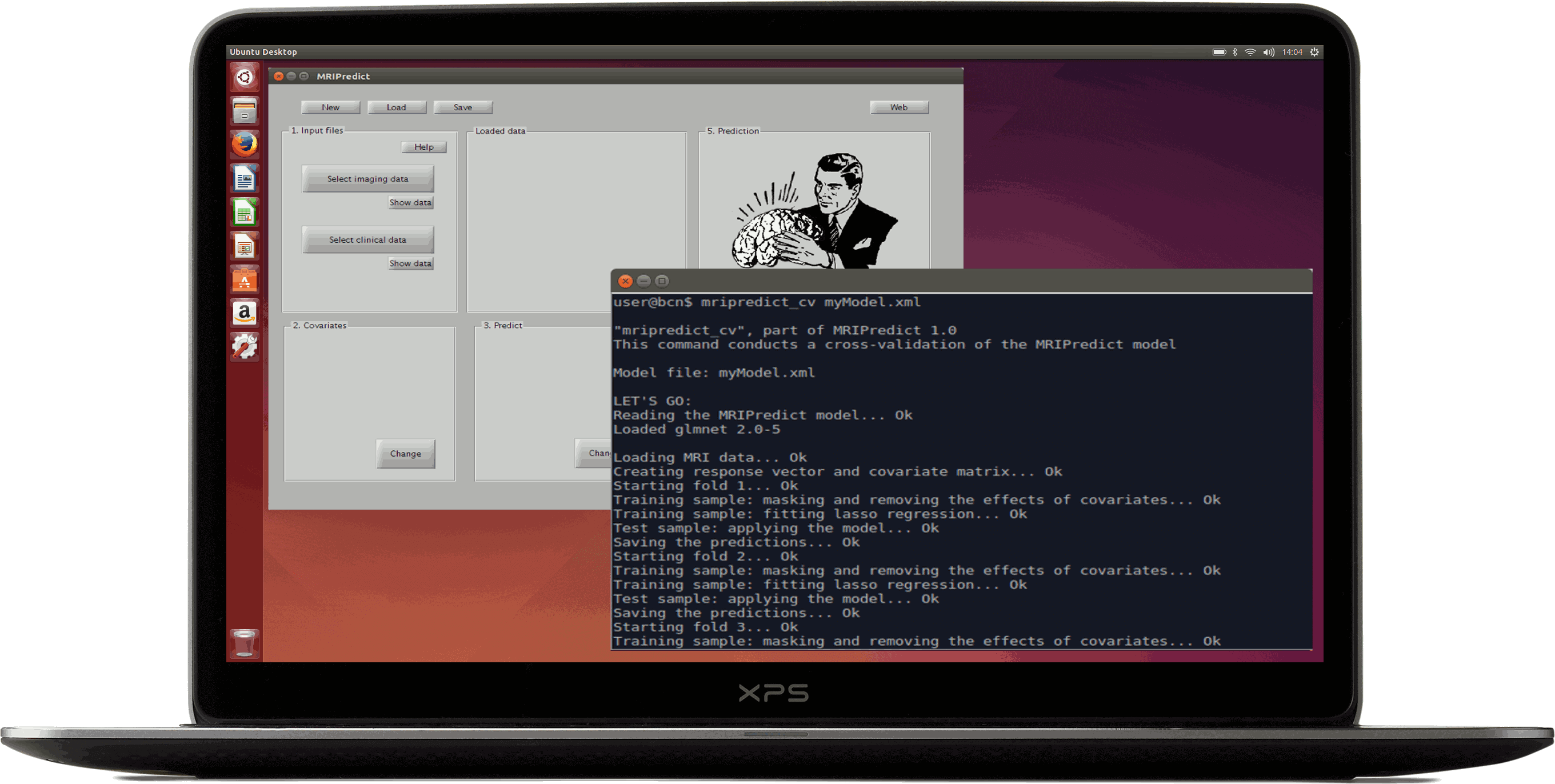
MRIPredict is a software package for using sMRI data to easily predict diagnosis (e.g. whether the patient will respond to a treatment, or whether the patient has one or another disorder).
Note that it is based on the glmnet library
The model may include covariates, and the software conducts both cross-validation of the model and fitting for its use with new VBM data.

Work co-funded by the Instituto de Salud Carlos III - Subdirección General de Evaluación y Fomento de la Investigación and the European Regional Development Fund (FEDER). Project grants CP14/00041, PI14/00292, PI14/01148 and PI14/01151.